- Home
- Who-is-Who
- Workshops
- Big Data in Electrophysiology
- Personalized health care workshop
- NeuroImaging Data
- Neuroinformatics congress 2015
- Atlasing workshop
- Neuroinformatics2014
- Cortical activity patterns
- Population Neuroimaging
- Image Analysis
- Imaging Summerschool Antwerp
- Predictive Neuroinformatics
- Neurotechnology Symposium
- Data sharing workshop
- Neural Systems modeling
- Donders Connectomes workshop
- Clinical Applications workshop
- Brain-inspired computing
- Dendrites workshop
- Share and Flourish
- Symposium 2011
- Calendar
- Companies
- Software & Services
- About / Mission
Atlas infrastructure and registration tools workshop
| Date: | Tu, December 2, 2014 |
| Time: | morning session 09:00 - 12:30, afternoon session 12:30-17:00 including lunch |
| Place: | Utrecht Pieterskerkhof 23 (route) |
| Registration: | Limited to 45 (morning), 25 (afternoon) participants.register here |
| Program committee: | Rembrandt Bakker, Fons Verbeek, Paul Tiesinga |
 |
 |
 |
An important mission of neuroinformatics is to promote new neuroscientific discoveries by providing an infrastructure in which brain-related data can be meaningfully shared and integrated. A major hurdle thereby is that brains differ across individuals, so that data measured at the same x,y,z-location in different brains cannot be compared. Brain atlasing is the scientific discipline that tackles this problem with two major strategies. The first is to try and attach data not to x,y,z-coordinates but to named brain regions that are defined by objective criteria, such as histochemical, electrophysiological and hodological (connectional) features. The second is to warp each brain in a dataset to a brain template that is chosen to be the standard. This warping, also called registration, can be surface- or volume based and optimizes some measure of similarity between the moving and fixed data. Warping is easiest when the two datasets have the same dimension (2d, 3d) and modality (T1, T2, Nissl, etc.), but the registration of slices to volumes and across modalities has also been successfully applied.
In this workshop, we look at progress that has been made in brain atlasing, with a particular emphasis on infrastructure and workflows that allow individual researchers to apply atlasing techniques to their own data. The workshop consists of two parts. In the morning session, atlasing experts present their work, tools and outlook. In the afternoon session, neuroscientists are invited to meet up with the atlasing experts to test and discuss whether the tools meet their requirements.
Program
| morning session (up to 50 participants) | ||
|---|---|---|
| 9:00 | Atlasing/registration tools session #1 | |
| Fons Verbeek (LIACS) | Building a digital atlasing infrastructure and studying zebrafish. | |
| Stefan Klein (EMC) and Marius Staring (LUMC) | Elastix software package for linear and nonlinear image registration. | |
| Rembrandt Bakker (Radboud Univ.) | Scalable brain atlas of mouse, rat, opossum, marmoset, macaque and human, with new registration services. | |
| Doug Bowden (Univ. Washington) | NeuroMaps slice to volume mapper, available for mouse and macaque. Tutorial (7MB pptx) | |
| Andrew Reid (FZ Jülich) | JuBrain Cytoarchitectonic Atlas applied to fMRI meta-analysis results. | |
| 10:30 | coffee/tea break | |
| 11:00 | Atlasing/registration tools session #2 | |
| Hakim Achterberg (EMC) | FASTR pipeline engine to accelerate imaging pipeline development. | |
| Boudewijn Lelieveldt | Pipeline to ingest the vast data resources of the Allen Institute. | |
| Trygve Leergaard and Jan G. Bjaalie | Infrastructure for digital atlasing of mouse and rat brain: Rodent Brain WorkBench |
|
| Nicole Schubert | Rat atlas with regional transmitter receptor distribution | |
| Stanislaw Adaszewski | Human brain project neuroinformatics platform: big data challenges in image registration. | |
| 12:30 | * * * end of morning session * * * | |
| afternoon session (up to 25 participants) | ||
| 12:30 | lunch in café Domkerk | |
| 13:30 | demos, tool feedback and use cases | |
| 15:30 | hackathon and collaborative work | |
| 17:00 | * * * end of afternoon session * * * | |
Route to Pieterskerkhof 23
View the route from Utrecht Central Station on Google maps. The distance is 1.2 km. If you lose your sense of direction: you have to walk past the big historical tower "Domtoren".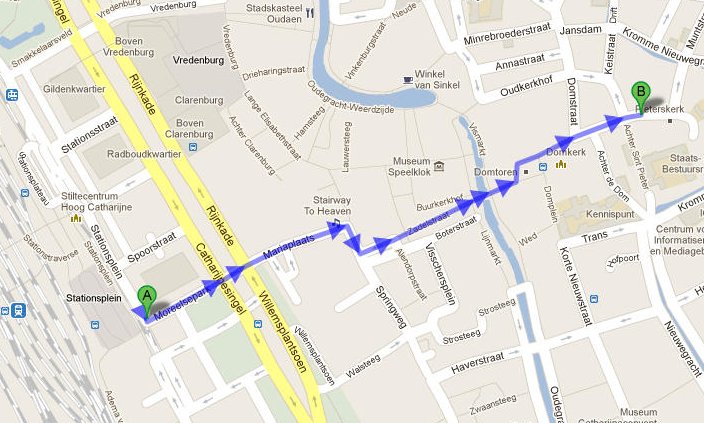
Date/period:
Tuesday, December 2, 2014 - 09:00
Location:
walking distance from Utrecht CS train station
Date status:
Fixed
Organizer:
NeuroInformatics.NL